Making upset plots is not something you do every day, admittedly. Upset plots are only useful in certain situations – but in those situations, they are entirely useful. It’s was hard for me to visualize a use-case, but let me list a few I have seen here:
- The most common demonstration I have seen of upset plots on the internet have been to try to characterize attributes of movies. This is probably because the creators of the package I use for upset plots, UpSetR, posted this example using movies with their documentation. They showed how some movies really have just one attribute – they are romance movies, or comedy movies. But then, where do you put the romantic comedy – the rom-com? That’s what the upset plot solves.
- My forever intern Natasha and I used it in a dashboard visualizing patterns of nosocomial infections in patients. Here, one patient can be infected with multiple organisms. We want to see what the patterns are of organism infection among the patients.
- Like we did with nosocomial data, the Gehlenborg Lab is doing with genomic data. See their short blog post about how they are making upset plots there.
To be honest, I did not find many good use-cases or examples of people using upset plots. I was analyzing data from the Behavioral Risk Factor Surveillance Survey (BRFSS) the other day, and I thought I’d share with you my upset plot.
Making Upset Plots for Co-morbidities
My colleague and I had a list of respondents from the BRFSS who had said “yes” to having one or more of the following co-morbidities: asthma, heart attack, coronary heart disease (CHD), stroke, chronic obstructive pulmonary disease (COPD), arthritis, depression, kidney disease, and/or diabetes. The “and/or” was the key. As soon as we got our sample, we wondered: Do these people all have a bunch of diseases? Or do most of them just have one disease? I recommended making upset plots to visualize these patterns.
Preparing the Data for UpSetR
The UpSetR package is actually quite flexible and can take in data in different formats. However, the easiest way I have found to use the package to get the upset plot I want is to prepare a dataset has this format:
- Every record (row) represents one experimental unit
- Each column is a yes/no flag as to whether or not they have the attribute, and is coded 1 for “yes” and 0 for “no”, and
- All the column names are the same as the names you want for the attribute on the plot.
I did this, and put it on Github for you here. The dataset is called Figure_Data.rds, and you will see that the code imports it, and then we can look at it.
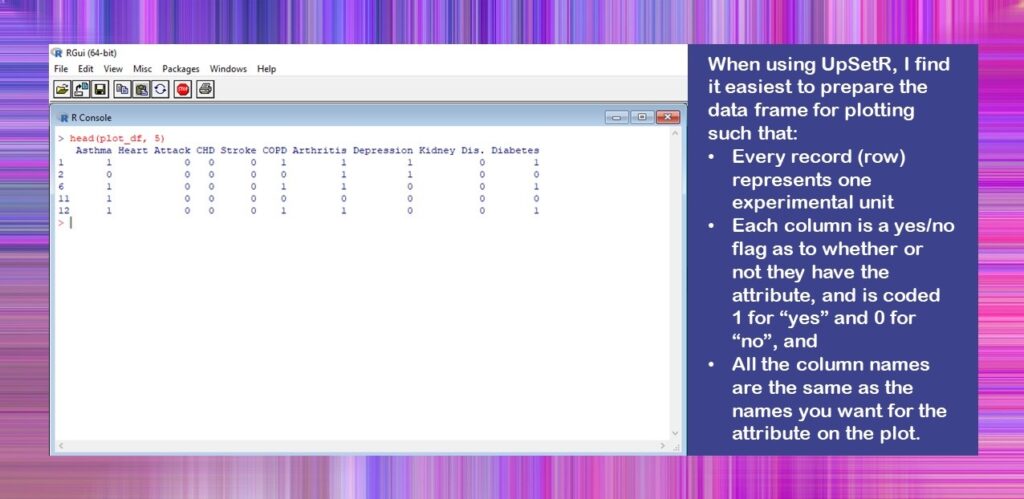
#read in analytic plot_df <- readRDS(file = "Figure_Data.rds") #look at colnames and what the data look like colnames(plot_df) head(plot_df, 5)
You will see I named our dataframe plot_df when I imported it into R. That’s because you usually have to create a special plot dataset in R when you want to plot something. This is different from SAS, where you usually just plot the whole dataset, and SAS figures it out for you (which is why it often runs a lot more slowly than R, but that’s another discussion altogether!).
You will also notice that instead of showing you the dataframe in Excel, I used the head() command to show the first five rows. This is because I was keeping this file as an RDS to improve data handling since it was such a large file, so it was never in Excel or *.csv format.
Here is what the console looks like when you run the head() code.
Setting Text Scale Options
The reason why setting text scale options is so challenging in the upset plot is because there is so much text. Please see below how I annotated my upset plot to show you the names of the text items that can be set in UpSetR. I also put a little “cheat sheet” on how you can make a vector (I called mine text_scale_options) with all these options set, which is what I did.
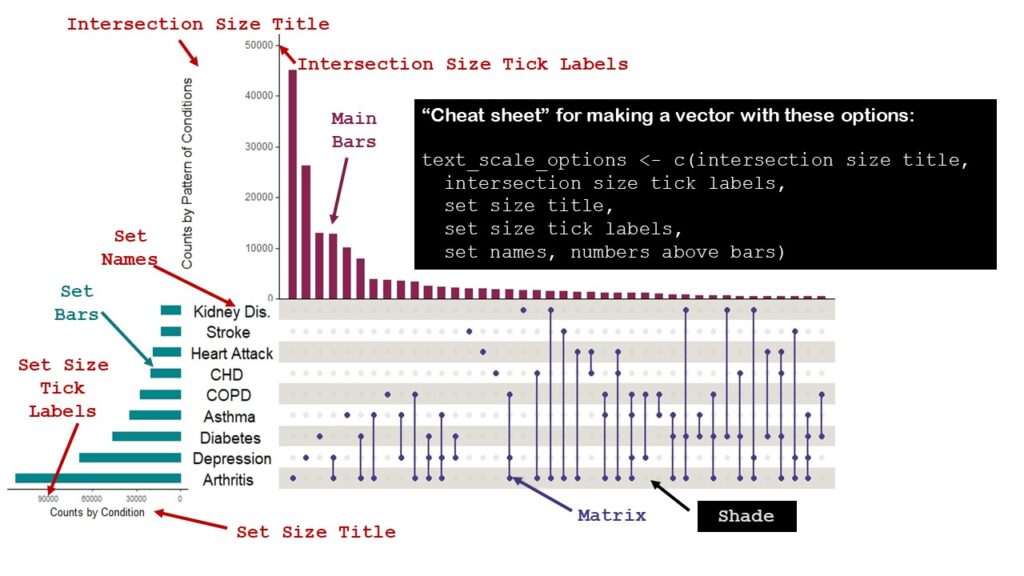
I did not choose to have data labels appear above the bars, but had I, could have set a text scale option for that, too. I wasn’t sure what options I’d choose, so I made a few sets, naming them 1, 2, and 3.
#I created different options so I could see which ones I liked best text_scale_options1 <- c(1, 1, 1, 1, 0.75, 1) text_scale_options2 <- c(1.3, 1.3, 1, 1, 2, 0.75) text_scale_options3 <- c(1.5, 1.25, 1.25, 1, 2, 1.5)
You will see when we get to the upset plot code that I end up using text_scale_options3.
Setting Colors
If you look at my graphic above, you will see that in addition to the text options, I named a few things that I colored: the main bars, the set bars, the matrix, and the shade. I also colored the labels on the graphic the same color as the items on the plot. That was to explain to you how I set the colors. I created four variables: main_bar_col, sets_bar_col, matrix_col, and shade_col, and set them to the colors I wanted for the main bars, the sets bars, the matrix, and the shade. Then, like with the text_scale_options3 vector, I used these variables when I ran the plot.
You can match the code below to the colors on the graphic.
#setting colors
#this can also be done with hexadecimal
main_bar_col <- c("violetred4")
sets_bar_col <- c("turquoise4")
matrix_col <- c("slateblue4")
shade_col <- c("wheat4")
Setting MB Ratio and Set Variables
To quote the authors from their documentation,
“To change the proportions of the plot heights assigned to the matrix and intersection size bar plot, use the mb.ratio parameter entered as percentages.”
I had a lot of trouble visualizing this. I found the best way to handle this was to create an mb.ratio vector (I called mb_ratio1), and then change it in my code, and make more vectors with different proportions if I didn’t like my first choices. As you will see by my code, I liked my first choices, so I only had one of them.
#I called it "1" because I anticipated others #but never needed them mb_ratio1 <- c(0.55,0.45)
Next, in the upset plot, you have to tell R which variables are the “set” variables – meaning which ones have your 1,0 flags in them. If you set your dataframe up as I recommended, then literally all of your variables are set variables.
In any case, again, it’s best to make a vector of the set variables. It’s important to note that the names of the columns in the dataframe will be the labels of your sets on the upset plot – so if you have weird variable names, you will want to change them before you make the plot.
- In that case, you could make a vector of plot-friendly variable names with the same number of entries as you have columns in your dataframe, and just use the names() command to rename all your columns.
- Also, if your dataframe already has the perfect set names, you could just use the colnames() command to create this vector.
In fact, I later realized that I could have done the second maneuver I described above – but I am satisfied with the way I have demonstrated it, because if you happen to have a dataframe with more variables in it than you want to plot, you would want to do it my way, which is just writing out the variable names in a character vector and saving it as set_vars.
set_vars <- c("Asthma", "Heart Attack", "CHD", "Stroke",
"COPD", "Arthritis", "Depression", "Kidney Dis.",
"Diabetes")
Making Upset Plots – Finally!
Before you make the plot, you need to make sure you install package UpSetR, and then run the library() command for UpSetR and ggplot2 (which I assume you have already installed since most R users have, but if not, you’ll need to install this one, too).
So here is the plot code:
upset(plot_df, sets = set_vars, mb.ratio = mb_ratio1, mainbar.y.label = "Counts by Pattern of Conditions", sets.x.label = "Counts by Condition", order.by = "freq", show.numbers = FALSE, point.size = 2, line.size = 1, text.scale=text_scale_options3, main.bar.color = main_bar_col, sets.bar.color = sets_bar_col, matrix.color = matrix_col, shade.color = shade_col)
And here is the plot without all that markup on it:
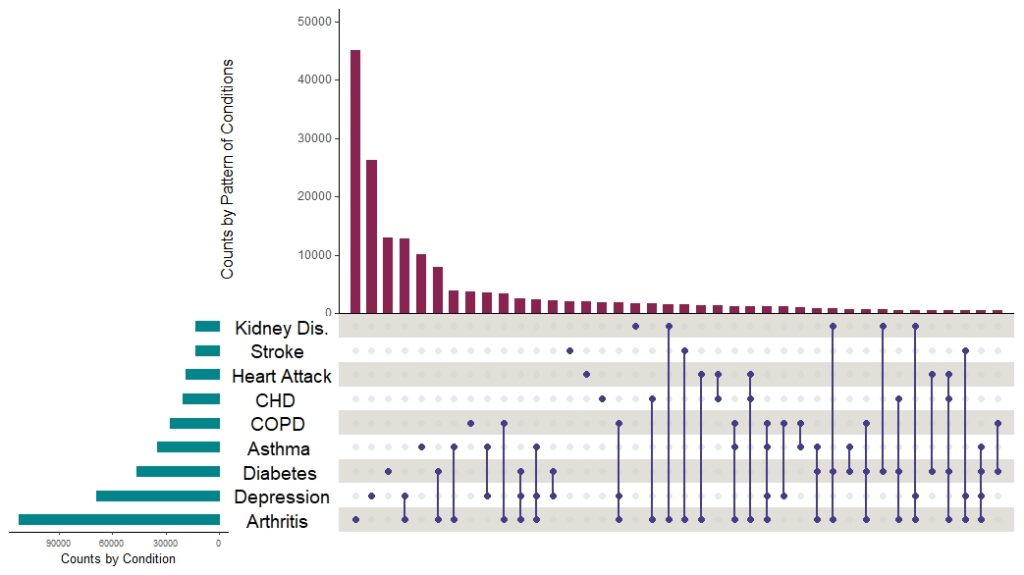
I want to point out a few things about making upset plots this way:
- First, please try to identify in the code where I used all the variables and vectors I set up. I find that is the best approach when making complex plots in R packages.
- You will see that I turned off the data labels with show.numbers = FALSE. That’s where you could turn them on, and set their text size in the options.
- You can see where I hardcoded labels in there. This is another place where you could put a vector, but I didn’t see myself fussing with these, so I hardcoded them.
- If you use this code to help you make an upset plot, and then you want to set other options I did not set, please read the documentation. I believe there are more ways to modify this plot, but the ones I presented are the ones I use most often.
As it turns out, the plot answered my question. The top three main bars were all singular conditions: arthritis, depression, and diabetes. The first pattern shows up as the fourth bar, and that one is arthritis plus depression. We found this visualization very helpful when we wrote our paper, because it demonstrated the distribution of conditions across our sample.
Published April 3, 2022. Added banners March 6, 2023. Revised banners June 19, 2023.
Read all of our data science blog posts!
Making upset plots with R package UpSetR is an easy way to visualize patterns of attributes in your data. My blog post demonstrates making patterns of co-morbidities in health survey respondents from the BRFSS, and walks you through setting text and color options in the code.





























































Hi Monika,
Thanks for the great and extensive explanation, it helped me a lot! In my dataset I have some cases that have missing values. So in your example, some individuals would have left one yes/no answer on the presence of a disease open. It know excludes these cases completely. Do you know if there is a way to still include the n/a’s?
Kind regards,
Frederique
Hi there,
Thank you for your comment! Yes, you are right. To restate what you are saying, I have reduced everyone to having a two-state flag – 1 or 0 – Yes or No. But what if the answer is “don’t know” – like, we don’t know if this person has arthritis because they never went to a physician and underwent a diagnostic visit?
Maybe you can see where I am going. I don’t think there is any value to including the prevalence or distribution of an unknown state in an upset plot. I have never gone for an arthritis diagnosis because I do not have any symptoms – but I’m pretty sure I don’t have it. People who go to a physician and undergo a diagnostic visit find out if they have it – yes or no – and then tell us when we ask in a survey.
So for this case, the two-state flag works. But if your measurement instrument is not that good – like, you are asking people what dishes they like from India, and they don’t know anything about Indian cuisine – well, you should probably rethink your measurement instrument. I guess that is where I’d finally come down on the question of graphing NA’s on an upset plot.
Hi Monika,
Thank you so much for this code! I’m having some trouble setting up an UpSet plot with it however. I’ve used your code exactly, except for importing my own data from excel in the format you recommended. But I get the following error:
Error in `data[, start_col:end_col]`:
! Can’t subset columns past the end.
i Location 22 doesn’t exist.
i There are only 21 columns.
Run `rlang::last_error()` to see where the error occurred.
Warning messages:
1: In xtfrm.data.frame(x) : cannot xtfrm data frames
2: In xtfrm.data.frame(x) : cannot xtfrm data frames
3: In xtfrm.data.frame(x) : cannot xtfrm data frames
4: In xtfrm.data.frame(x) : cannot xtfrm data frames
5: In xtfrm.data.frame(x) : cannot xtfrm data frames
Do you have any idea how to solve this?
Hello dear,
Well, it’s hard to troubleshoot in the comments, but I’ll try. I see “Error in ‘data[,start_col:end_col]'” which suggests that there is some error reading in your data. I had a boss who taught me to “Google your error messages” so when I Googled “In xtfrm.data.frame(x) : cannot xtfrm data frames” (because I see that being repeated) and I find a few different causes of this error. I think the most revealing part of the error is “Location 22 doesn’t exist – there are only 21 columns”. You may need to regenerate your dataframe (I get this from reading the different doco about your error message). Here is one way – to make a matrix first then convert it to a dataframe: https://youtu.be/E8wl_P6yZkw Sorry I couldn’t be of more help, but feel free to provide more info, and I can try again. Hopefully a little messing around with code fixes it. Good luck!
-Monika
Hey Monika,
The dataset seems so interesting. Can you send it to me by email? I am a PhD student at the University of Utah working with UpSet Plot’s inventor! We are currently collecting data for research purposes.
Regards,
Ishrat Jahan Eliza
Visualization Design Lab
Hi, I am new to using R. What sort of data can be represented on an UpSet Plot? Can you please simplify? I work with NGS data—variants detected in solid tumours. I cannot find this specific example anywhere in the making of and UpSet plot. Thank you