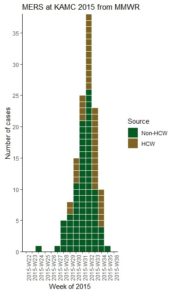
With all this talk about “flattening the curve” of the coronavirus, I thought I would get into the weeds about what curve we are talking about when we say that. We are talking about what’s called an epidemiologic curve, or epicurve for short. And to demonstrate what an epicurve is and what it means, I used the R package EpiCurve to demonstrate making an epidemiologic curve so you can see how it works.
Source Data for my Epicurve
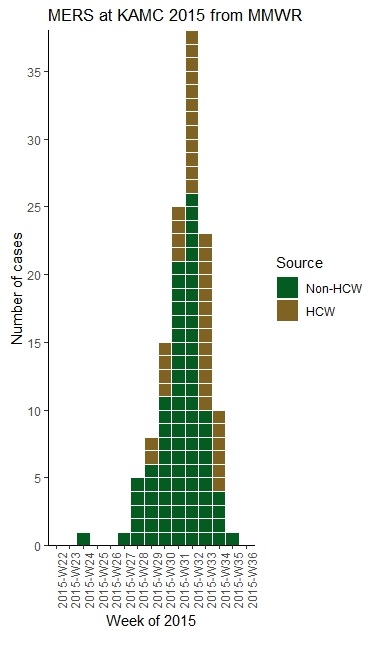
I actually back-engineered the data for my dataset based on viewing the epicurve in this article from Morbidity and Mortality Weekly Report of a nosocomial outbreak of Middle East respiratory syndrome (MERS) at King Abdulaziz Medical City in Riyadh, Saudi Arabia in June through August, 2015.
The downside to back-engineering data is that it might have errors, but the advantage is that I can assemble it into the perfect structure for the R package EpiCurve. If you look in the article, they have an epicurve, but they don’t include the raw numbers, so I just guessed the data. The data I guessed are below.
| RowID | Start_of_Week | End_of_Week | Frequency | Source |
| 1 | 2015-05-31 | 2015-06-06 | 0 | Non-HCW |
| 2 | 2015-06-07 | 2015-06-13 | 0 | Non-HCW |
| 3 | 2015-06-14 | 2015-06-20 | 1 | Non-HCW |
| 4 | 2015-06-21 | 2015-06-27 | 0 | Non-HCW |
| 5 | 2015-06-28 | 2015-07-04 | 0 | Non-HCW |
| 6 | 2015-07-05 | 2015-07-11 | 1 | Non-HCW |
| 7 | 2015-07-12 | 2015-07-18 | 5 | Non-HCW |
| 8 | 2015-07-19 | 2015-07-25 | 2 | HCW |
| 9 | 2015-07-19 | 2015-07-25 | 6 | Non-HCW |
| 10 | 2015-07-26 | 2015-08-01 | 4 | HCW |
| 11 | 2015-07-26 | 2015-08-01 | 11 | Non-HCW |
| 12 | 2015-08-02 | 2015-08-08 | 4 | HCW |
| 13 | 2015-08-02 | 2015-08-08 | 21 | Non-HCW |
| 14 | 2015-08-09 | 2015-08-15 | 12 | HCW |
| 15 | 2015-08-09 | 2015-08-15 | 26 | Non-HCW |
| 16 | 2015-08-16 | 2015-08-22 | 13 | HCW |
| 17 | 2015-08-16 | 2015-08-22 | 10 | Non-HCW |
| 18 | 2015-08-23 | 2015-08-29 | 6 | HCW |
| 19 | 2015-08-23 | 2015-08-29 | 4 | Non-HCW |
| 20 | 2015-08-30 | 2015-09-05 | 0 | HCW |
| 21 | 2015-08-30 | 2015-09-05 | 1 | Non-HCW |
| 22 | 2015-09-06 | 2015-09-12 | 0 | HCW |
| 23 | 2015-09-06 | 2015-09-12 | 0 | Non-HCW |
Variables for the R Package EpiCurve
Let’s look more closely at each variable:
- RowID: This is just the number of the row. I always include this as a unique identifier in tables.
- Start_of_Week: The epicurve in the article presents each frequency bar as representing a week. This variable is the day at the start of the week.
- End_of_Week: This is the day at the end of the week.
- Frequency: This is the number of cases for the week from the specified Source (see next column).
- Source: You will see in the epicurve in the article, each bar is color-coded for healthcare worker (HCW) or non-HCW. To facilitate this color coding, each category needs its own row. You will see in the first rows, there were no cases, so I just put one row, randomly assigned it a source, and put 0. In rows 6 and 7, only non-HCW’s were cases, so those weeks have only one row. Whenever there were HCW and non-HCW identified as cases in the same week, they were each expressed in their own row.
Programming for the R EpiCurve Package
First, I knew I’d need two colors – one for HCW and one for non-HCW so I used the Coolors app and the logo for the Saudi Arabia Ministry of Health to pick my colors in hexadecimal. Once I selected them, I loaded them into variables, one called saudi_gold and one called saudi_green.
saudi_gold <- c("#806222")
saudi_green <- c("#045c20")
That way, I could just refer to them as saudi_gold and saudi_green when programming.
Next, after installing the EpiCurve package, I called up the library, and made these specifications to get the epicurve shown at the top the post.
library(EpiCurve)
EpiCurve(MERS,
date= "Start_of_Week",
period = "day",
to.period = "week",
freq = "Frequency",
cutvar = "Source",
colors = c(saudi_gold, saudi_green),
title = c("MERS at KAMC 2015 from MMWR"),
ylabel = c("Number of cases"),
xlabel = c("Week of 2015"))
Here are some important parts of the code above:
- The EpiCurve package handles dates in factor format! In fact, converting them to date won’t work. So as you can see in the code above, where
date=”Start_of_Week”, that date variable – as well as the date variable End_of_Week – are both actually in factor format.
- Period and to.period: These had be correct in order for the date frequency to collapse and come out right as weeks. Many epicurves are by days, so if
period=day and the data are daily so they do not have to be collapsed into weeks or months, the to.period argument can be left out.
- Cutvar is the command to specify the categories: The argument
curtvar=”Source” is what facilitated the epicurve looking at the categorical column Source that separates HCW and Non-HCW data.
- Colors are specified using the variables set: This makes it easier to see what colors are being specified. You will see that between
cutvar and the colors specification, the legend is automatically output.
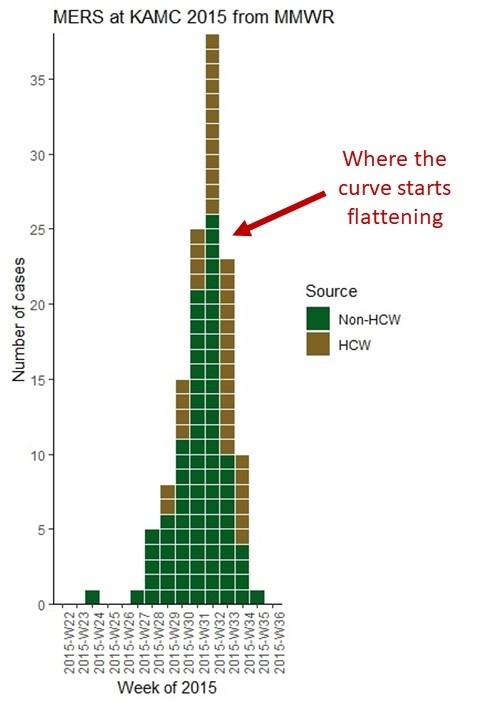
Where the Epicurve Starts Flattening
The reason we make an epicurve during an epidemic is that in addition to tracking the mortality rate, we are interested in tracking when the curve actually begins to flatten. Note where I annotated the epicurve from the MERS article to point out the week that that curve finally flattened. When the curve finally flattens, you are literally “on the other side” of the outbreak. The problem is that during the epidemic you never really know you are there until you get there, and the day or week you are in finally sees fewer new cases than the day or week before.
As you proceed through the epidemic, as long as you keep formatting your data the way I demonstrated, you can keep adding to it and generating a new epicurve every day.
Read all of our data science blog posts!
Confidence intervals (CIs) help you get a solid estimate for the true population measure. Read [...]
Continuous variable categorized can open up a world of possibilities for analysis. Read about it [...]
Delete if rows meet a certain criteria is a common approach to paring down a [...]
Chi-square test is hard to grasp – but doing it in Microsoft Excel can give [...]
Identify elements in research reports, and you’ll be able to understand them much more easily. [...]
Time periods are important when creating a time series visualization that actually speaks to you! [...]
Apply weights to get weighted proportions and counts! Read my blog post to learn how [...]
Make categorical variables by cutting up continuous ones. But where to put the boundaries? Get [...]
Remove rows by criteria is a common ETL operation – and my blog post shows [...]
CDC Wonder is an online query portal that serves as a gateway to many government [...]
AI careers are not easy to navigate. Read my blog post for foolproof advice for [...]
Descriptive analysis of Black Friday Death Count Database provides an example of how creative classification [...]
Classification crosswalks are easy to make, and can help you reduce cardinality in categorical variables, [...]
FAERS data are like any post-market surveillance pharmacy data – notoriously messy. But if you [...]
Dataset source documentation is good to keep when you are doing an analysis with data [...]
Joins in base R must be executed properly or you will lose data. Read my [...]
NHANES data piqued your interest? It’s not all sunshine and roses. Read my blog post [...]
Color in visualizations of data curation and other data science documentation can be used to [...]
Defaults in PowerPoint are set up for slides – not data visualizations. Read my blog [...]
Text and arrows in dataviz, if used wisely, can help your audience understand something very [...]
Shapes and images in dataviz, if chosen wisely, can greatly enhance the communicative value of [...]
Table editing in R is easier than in SAS, because you can refer to columns, [...]
R for logistic regression in health data analytics is a reasonable choice, if you know [...]
Connecting SAS to other applications is often necessary, and there are many ways to do [...]
Portfolio project examples are sometimes needed for newbies in data science who are looking to [...]
Project management terminology is often used around epidemiologists, biostatisticians, and health data scientists, and it’s [...]
“Rapid application development” (RAD) refers to an approach to designing and developing computer applications. In [...]
Understanding legacy data is necessary if you want to analyze datasets that are extracted from [...]
Front-end decisions are made when applications are designed. They are even made when you design [...]
Reducing query cost is especially important in SAS – but do you know how to [...]
Curated datasets are useful to know about if you want to do a data science [...]
Statistics trivia for data scientists will refresh your memory from the courses you’ve taken – [...]
Management tips for data scientists can be used by anyone – at work and in [...]
REDCap mess happens often in research shops, and it’s an analysis showstopper! Read my blog [...]
GitHub beginners – even in data science – often feel intimidated when starting their GitHub [...]
ETL pipeline documentation is great for team communication as well as data stewardship! Read my [...]
Benchmarking runtime is different in SAS compared to other programs, where you have to request [...]
End-to-end AI pipelines are being created routinely in industry, and one complaint is that academics [...]
Referring to columns in R can be done using both number and field name syntax. [...]
The paste command in R is used to concatenate strings. You can leverage the paste [...]
Recoloring plots in R? Want to learn how to use an image to inspire R [...]
Adding error bars to ggplot2 in R plots is easiest if you include the width [...]
“AI on the edge” was a new term for me that I learned from Marc [...]
Pie chart ggplot style is surprisingly hard to make, mainly because ggplot2 did not give [...]
Time series plots in R are totally customizable using the ggplot2 package, and can come [...]
Data curation solution that I posted recently with my blog post showing how to do [...]
Making upset plots with R package UpSetR is an easy way to visualize patterns of [...]
Making box plots in R affords you many different approaches and features. My blog post [...]
Convert CSV to RDS is what you want to do if you are working with [...]
GPower case example shows a use-case where we needed to select an outcome measure for [...]
Querying the GHDx database is challenging because of its difficult user interface, but mastering it [...]
Variable names in SAS and R are subject to different “rules and regulations”, and these [...]
Referring to variables in processing is different conceptually when thinking about SAS compared to R. [...]
Counting rows in SAS and R is approached differently, because the two programs process data [...]
Native formats in SAS and R of data objects have different qualities – and there [...]
Looking for a SAS-R integration example that uses the best of both worlds? I show [...]
Want to compare multiple rankings on two competing items – like hotels, restaurants, or colleges? [...]
Getting data for meta-analysis together can be challenging, so I walk you through the simple [...]
Get to know three of my favorite SAS documentation pages: the one with sort order, [...]
I use the datasets from the Behavioral Risk Factor Surveillance Survey (BRFSS) to demonstrate in [...]
I love the Likert package in R, and use it often to visualize data. The [...]
With all this talk about “flattening the curve” of the coronavirus, I thought I would [...]
During my failed attempt to get a PhD from the University of South Florida, my [...]
At the beginning of the pandemic, I used the EpiCurve package in R to make an early epicurve. I show you my code in this blog post.